Table Of Content
- Design of Monomeric Water-Soluble β-Hairpin and β-Sheet Peptides
- Sequence design using machine learning methods
- Extended Data Fig. 1 Training ablations reveal determinants of RFdiffusion success.
- Enhancing the pharmaceutical properties of protein drugs by ancestral sequence reconstruction
- Opportunities and challenges in design and optimization of protein function
- Data availability
- Computational Redesign of Metalloenzymes for Catalyzing New Reactions

We used RFdiffusion to design C3-symmetric trimers that rigidly hold three binding domains (the functional motif in this case) such that they exactly match the ACE2 binding sites on the SARS-CoV-2 spike protein trimer. The designs were confidently predicted by AF2 to both assemble as C3-symmetric oligomers, and to scaffold the AHB2 SARS-CoV-2 binder interface with high accuracy (Fig. 5a). Chemistry plays a vital role in protein design by helping researchers understand the forces and interactions that govern protein folding. Protein folding refers to the process by which a linear chain of amino acids folds into a three-dimensional structure, which determines its function. Understanding the rules of protein folding allows us to design proteins that adopt their intended structures and exhibit desired functions. These sub-disciplines involve the development and application of algorithms and software tools to analyze biological data, enabling researchers to predict protein structures, identify potential target sites for modification, and model the behavior of designed proteins.
Design of Monomeric Water-Soluble β-Hairpin and β-Sheet Peptides
A pretrained deep convolutional neural network then recovers the three-dimensional backbone structure from pairwise distances. Some of the designed structures could be recapitulated by fragment-based structure prediction methods (82). Another variational autoencoder–based model focused on generating immunoglobulin structures (83). The model learned the distribution of immunoglobulin structures and compressed the distribution into a low-dimensional space termed latent space.
Sequence design using machine learning methods
A high-resolution cryo-EM structure of one of these designs in complex with influenza HA shows that RFdiffusion can design functional proteins with atomic accuracy. Vázquez Torres et al. demonstrate the ability of RFdiffusion to design picomolar affinity binders to flexible helical peptides54, further highlighting its use for de novo binder design. Vázquez Torres et al. also show how RFdiffusion can be extended for protein model refinement by partial noising and denoising, which enables tuneable sampling around a given input structure. For peptide binder design, this enabled increases in affinity of nearly three orders of magnitude without high-throughput screening. Despite not being trained on symmetric inputs, RFdiffusion is able to generate symmetric oligomers with high in silico success rates (Extended Data Fig. 5b), particularly when guided by an auxiliary inter- and intrachain contact potential (Extended Data Fig. 5c).
De novo design of modular protein hydrogels with programmable intra- and extracellular viscoelasticity Proceedings ... - pnas.org
De novo design of modular protein hydrogels with programmable intra- and extracellular viscoelasticity Proceedings ....
Posted: Tue, 30 Jan 2024 18:50:41 GMT [source]
Extended Data Fig. 1 Training ablations reveal determinants of RFdiffusion success.
Given an input backbone structure, SPDesign utilizes ultrafast shape recognition vectors to accelerate the search for similar protein structures in our in-house PAcluster80 structure database and then extracts the sequence profile through structure alignment. Combined with structural pre-trained knowledge and geometric features, they are further fed into an enhanced graph neural network for sequence prediction. The results show that SPDesign significantly outperforms the state-of-the-art methods, such as ProteinMPNN, Pifold and LM-Design, leading to 21.89%, 15.54% and 11.4% accuracy gains in sequence recovery rate on CATH 4.2 benchmark, respectively.
Enhancing the pharmaceutical properties of protein drugs by ancestral sequence reconstruction
By leveraging the power of computational modeling, our researchers are creating solutions to some of the most pressing challenges in medicine, technology, and sustainability. In this post, we will delve into the basics of protein design, explore its real-world applications, and learn about the interdisciplinary nature of this fascinating field. In September, Redmond, Wash.-based Microsoft released an open-source model to generate proteins, and other big tech companies are also betting on the field. Straightaway the AI designed new molecules that also increase the activity of PPARs, like the drugs currently available, but without a lengthy discovery process.
Libraries of potential protein–protein interacting partners can be screened by this assay. An advantage of the split GFP assay is that protein-binding partners can be examined in the context of their native cellular environment, whether it be E. The precise order of amino acids in a protein chain, like the letters in a word, is crucial. The unique functions of a protein stem from its three-dimensional structure, which is determined by the sequence of amino acids composing the protein. Protein engineers are drawing on rapidly evolving machine learning tools, deep reservoirs of data, and the structure-predicting firepower of AlphaFold2 to pursue more sophisticated de novo protein designs.
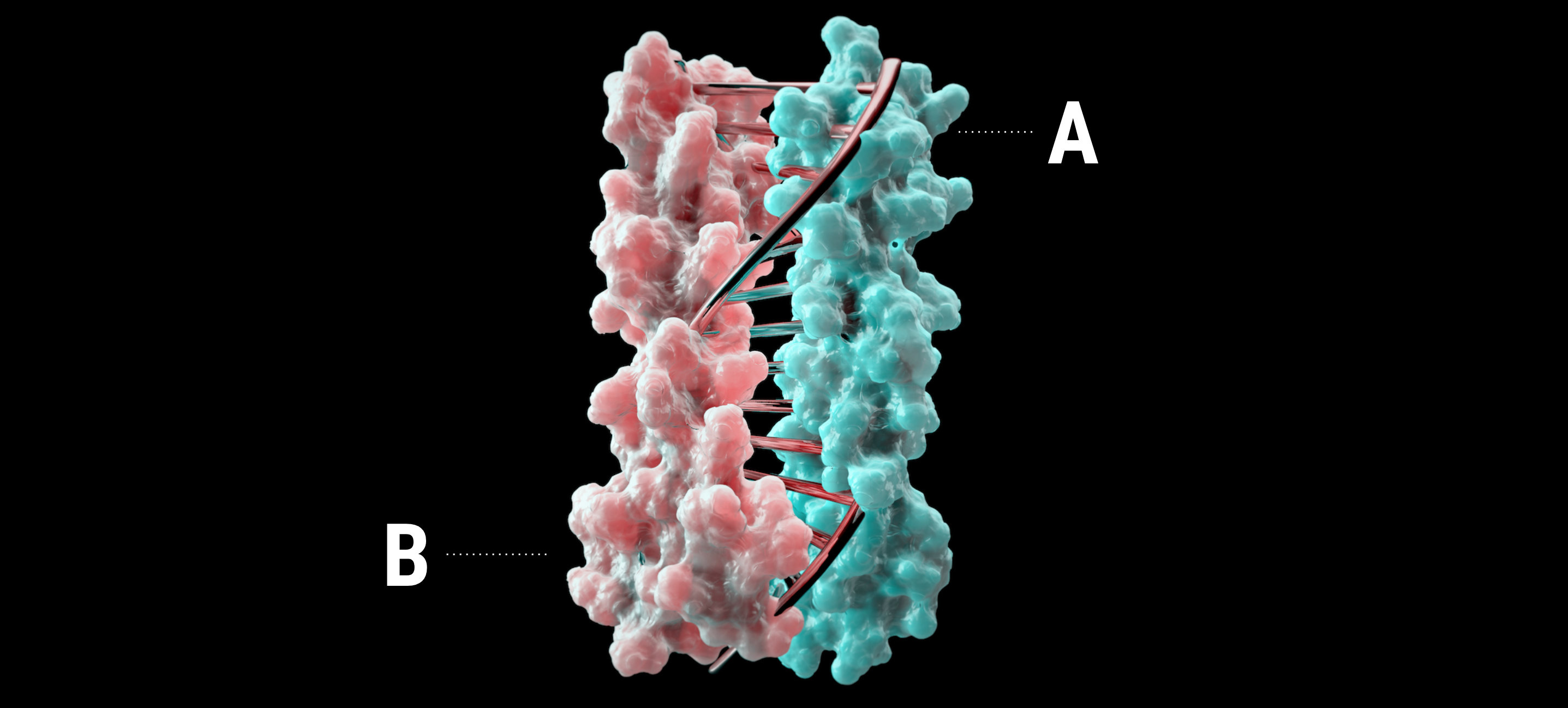
Like the SAGE particles vide supra, the large cavities in these protein assemblies have potential applications in delivery of molecular cargo. A major breakthrough in protein design that allows easy expression of branched protein building blocks was made by Howarth and colleagues. Protein expression is typically limited to linear topologies, but the development of SpyCatcher/Spy-Tag technology has changed that. SpyTag and SpyCatcher are peptide and protein components, respectively, that originated from splitting a fibronectin-binding domain of a bacterial adhesin. This fibronectin-binding domain spontaneously forms an intramolecular covalent bond in nature, and researchers were able to maintain this activity whilst dividing the protein into two separate parts49 (Figure 9, panel A).
Figures
Algorithm designs proteins from scratch that can bind drugs and small molecules - Chemistry World
Algorithm designs proteins from scratch that can bind drugs and small molecules.
Posted: Fri, 12 Apr 2024 07:00:00 GMT [source]
Find and hire a designer to make your vision come to life, or host a design contest and get ideas from designers around the world. A client requested a new packaging design for their Whey Protein product, to coincide with an upcoming rebranding and label update. The design specifications required it to be visually appealing to a broad audience, utilize legible fonts, and be suitable for online sales. To achieve this, I used a dark neutral base with embossed textures in certain areas to provide a premium user experience. The accent brown-copper gradient color was chosen to represent the chocolate flavor of this particular variant.
A back-rub motion involves internal backbone rotations about axes between C-alpha atoms. Incorporating such back-rub moves into design simulations has led to considerable improvements in modeling structural changes in point mutants (49, 50, 52), protein dynamics on fast timescales (53, 54), prediction of molecular recognition specificity (55), and the sequence design (56). Gene editing has the potential to solve fundamental challenges in agriculture, biotechnology, and human health. CRISPR-based gene editors derived from microbes, while powerful, often show significant functional tradeoffs when ported into non-native environments, such as human cells.
It will also be useful to develop molecular switches that can respond to biologically relevant stimuli. These would greatly aid the creation of synthetic circuits for therapeutic, industrial, and detection applications. Another major focus will be to create new methods for controlling proteins with light. Light-sensitive domains will be developed that can control a wider range of protein activities, respond to light stimuli faster, and be induced by longer wavelengths than existing designs. The future of designed materials and assemblies lies in the creation of more diverse and robust protein building blocks. The development of new types of specific protein–protein interaction modules with minimal cross-reactivity with each other or with cellular components would also be useful.100 Additionally, new and improved “smart” materials will focus on enhanced responsiveness to molecular stimuli.
Recently, a new membrane scoring model (124) was developed, which aims to better capture the heterogeneous membrane environment (Fig. 4A). The interface between bulk water and bulk lipid is modeled as a continuous transition of hydration fraction, with water-filled pores modeled using a convex-hull algorithm (125). The water-to-bilayer transfer energy is then calculated using the hydration fraction and the Moon and Fleming hydrophobicity scale (126).
Recently, the TERM score was used to predict protein–peptide binding energies and design peptide binders of antiapoptotic proteins Bfl-1 and Mcl-1 (134). Scoring functions for soluble proteins take advantage of the large number of solved structures in the PDB to validate and fit the parameters of the score function (121, 122). Transmembrane proteins make up about 30% of ORFs in known genomes but are currently underrepresented in the PDB, complicating the development of membrane protein scoring functions. An early version of the Rosetta membrane scoring function (123) used statistics from 28 transmembrane proteins to fit parameters and was validated by ab initio structure prediction of 12 multipass membrane proteins.
Several hundred high-affinity binders were validated by a high-throughput yeast surface display assay. Likewise, proteins that bind to the interleukin-2 and interleukin-15 receptors were designed by building a helical bundle from interface helices of native interleukin-2 and interleukin-15 (6). De novo backbones beyond helical bundles can be designed by a fragment assembly strategy originally used in structure prediction (35, 60). Typically, the first step in design is defining a blueprint that specifies the lengths and relative orientations of secondary structure elements.

No comments:
Post a Comment